Overview
This vignette shows how to: 1) simulate an epidemic using epiworldR , andcalibrate_sir()epiworldRcalibrate .
✅ No Python setup required. The package initializes the Python model internally the first time you call calibrate_sir() and cleans up automatically when asked.
Libraries
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 4.0.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.4
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library (ggplot2)library (patchwork)library (epiworldR)
Thank you for using epiworldR! Please consider citing it in your work.
You can find the citation information by running
citation("epiworldR")
Attaching package: 'epiworldR'
The following object is masked from 'package:lubridate':
today
library (epiworldRcalibrate)
Ground-Truth Parameters and Simulation
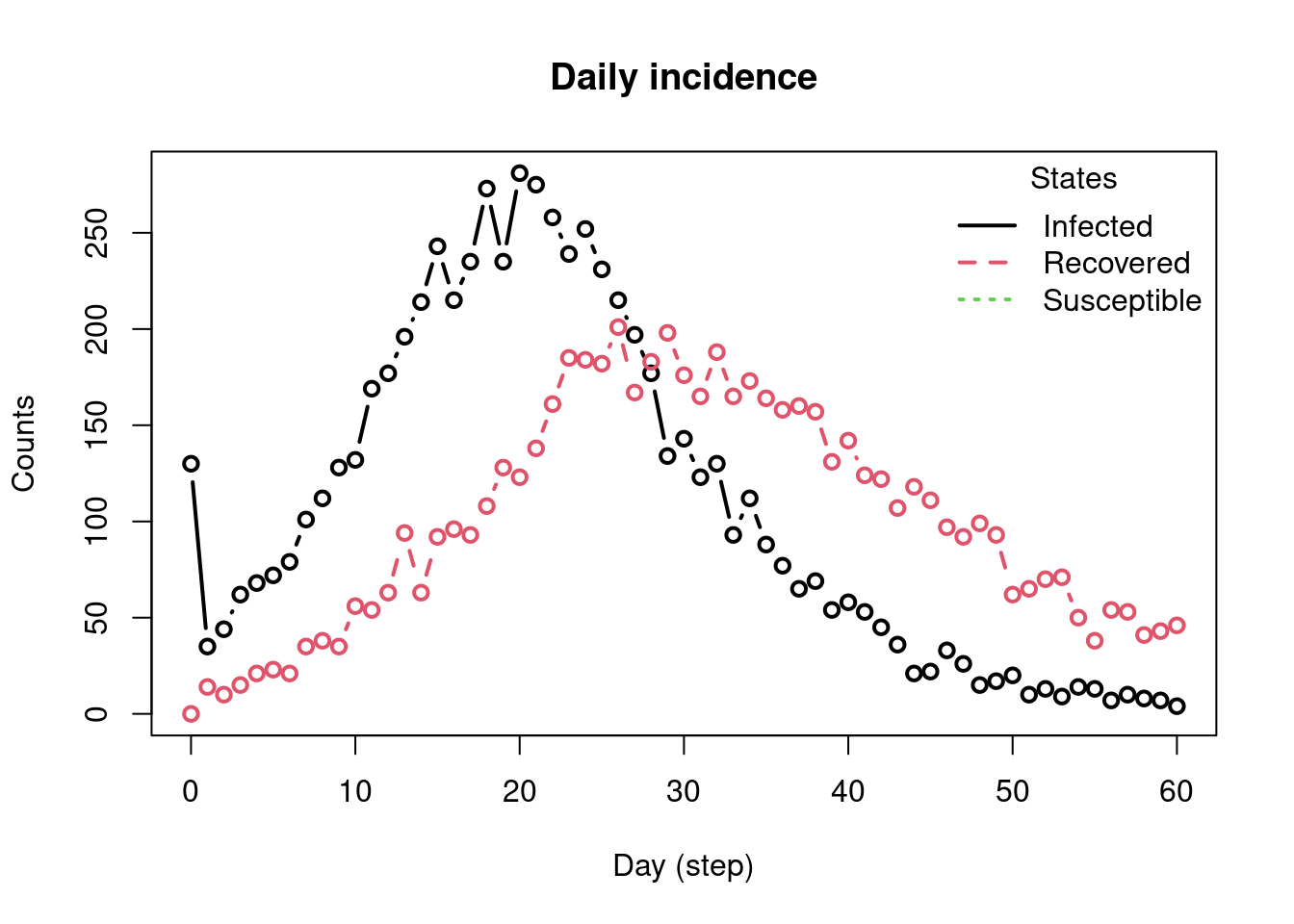
We draw a single SIR parameter set and simulate 60 days.
set.seed (122 )# Draw a single parameter set <- sample (5000 : 10000 , 1 )<- runif (1 , 0.007 , 0.02 )<- runif (1 , 1 , 5 )<- runif (1 , 0.071 , 0.25 )<- runif (1 , 1.1 , 5 )<- R0_true * recov / crate<- tibble (n = n_value,prevalence = preval,contact_rate = crate,transmission_rate = ptran,recovery_rate = recov,R0 = R0_true
# A tibble: 1 × 6
n prevalence contact_rate transmission_rate recovery_rate R0
<int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 6967 0.0188 1.76 0.149 0.0783 3.36
<- 60 <- ModelSIRCONN (name = "true_simulation" ,n = true_params$ n,prevalence = true_params$ prevalence,contact_rate = true_params$ contact_rate,transmission_rate = true_params$ transmission_rate,recovery_rate = true_params$ recovery_raterun (true_model, ndays = ndays)
_________________________________________________________________________
|Running the model...
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.
|
# Extract daily incidence (length must be 61: day 0..60) <- plot_incidence (true_model, plot = TRUE )<- inc_plot[, 1 ]length (incidence_ts) # should be 61
Calibrate SIR Parameters (One line)
calibrate_sir() automatically:
initializes the BiLSTM model (if not already loaded),
preprocesses the series,
predicts ptran , crate , and R0 ,
and returns a named vector.
<- calibrate_sir (daily_cases = incidence_ts,population_size = true_params$ n,recovery_rate = true_params$ recovery_rate
Using existing virtual environment: epiworldRcalibrate
Verifying package installation...
✓ numpy v2.2.6 loaded successfully
✓ sklearn v1.7.2 loaded successfully
✓ joblib v1.5.2 loaded successfully
✓ torch v2.9.1+cpu loaded successfully
All required Python packages are ready!
BiLSTM model loaded successfully.
cat ("LSTM Parameter Predictions: \n " )
LSTM Parameter Predictions:
ptran crate R0
0.1087724 2.4655740 3.4240935
Turn predictions into a tidy frame for comparison:
<- tibble (n = true_params$ n,prevalence = true_params$ prevalence,contact_rate = lstm_predictions[["crate" ]],transmission_rate = lstm_predictions[["ptran" ]],recovery_rate = true_params$ recovery_rate,R0 = lstm_predictions[["R0" ]]<- bind_rows (%>% mutate (param_type = "true" ),%>% mutate (param_type = "lstm" )
# A tibble: 2 × 7
n prevalence contact_rate transmission_rate recovery_rate R0 param_type
<int> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 6967 0.0188 1.76 0.149 0.0783 3.36 true
2 6967 0.0188 2.47 0.109 0.0783 3.42 lstm
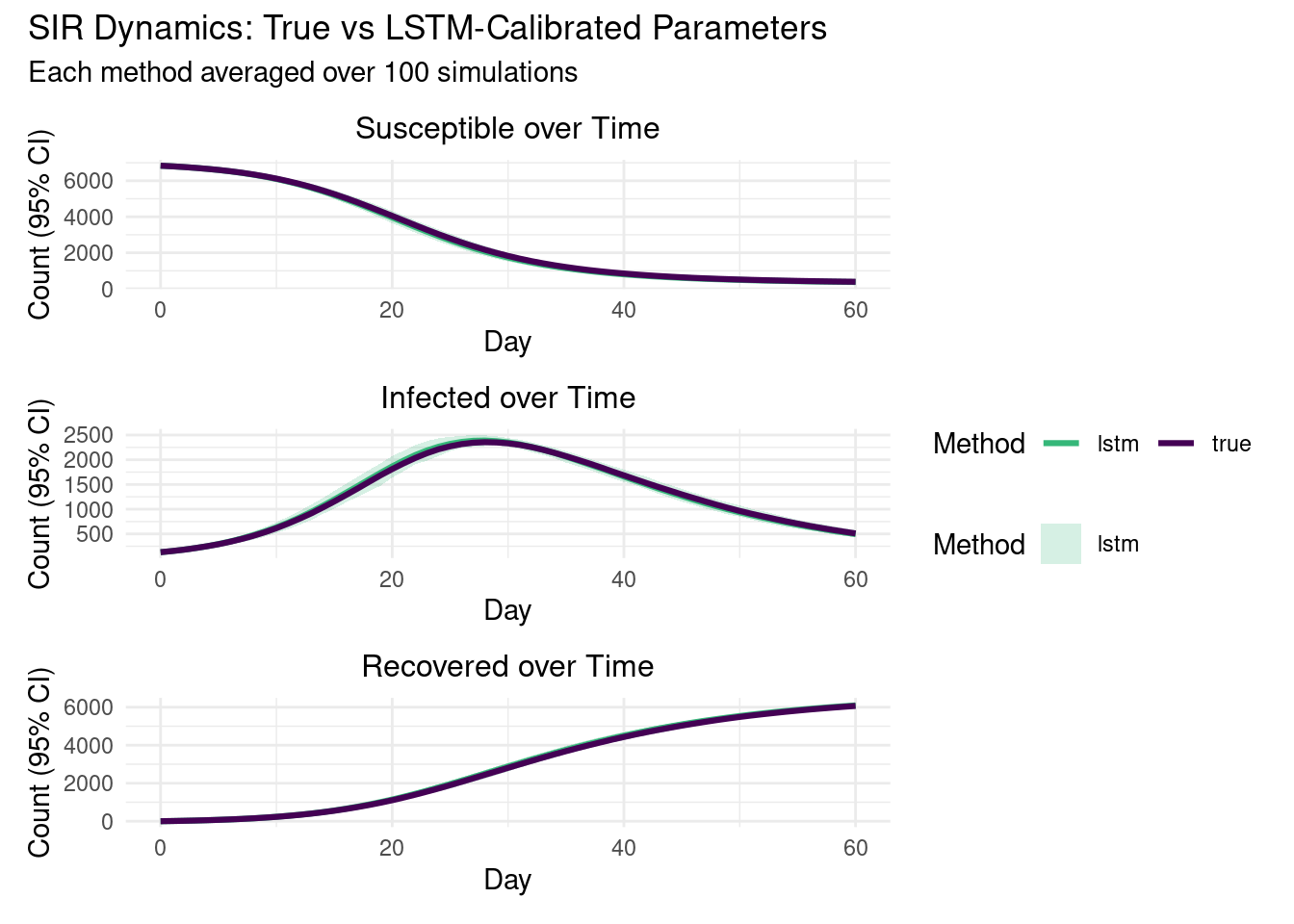
Forward Simulations: True vs LSTM Parameters
We run multiple replicates with the true parameters and with the LSTM-calibrated parameters to compare dynamics.
<- 100 <- tibble ()for (i in seq_len (nrow (params_comparison))) {<- params_comparison[i, ]<- ModelSIRCONN (name = paste0 ("forward_" , row$ param_type),n = row$ n,prevalence = row$ prevalence,contact_rate = row$ contact_rate,transmission_rate = row$ transmission_rate,recovery_rate = row$ recovery_rate<- make_saver ("total_hist" )run_multiple (forward_model, ndays = ndays, nsims = n_reps, saver = saver, nthreads = 8 )<- run_multiple_get_results (forward_model, nthreads = 2 )<- results$ total_hist %>% group_by (date, state) %>% summarize (mean_count = mean (counts),ci_lower = quantile (counts, 0.025 ),ci_upper = quantile (counts, 0.975 ),.groups = "drop" %>% mutate (param_type = row$ param_type)<- bind_rows (all_simulation_results, sim_data)
Starting multiple runs (100) using 8 thread(s)
_________________________________________________________________________
_________________________________________________________________________
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.
Starting multiple runs (100) using 8 thread(s)
_________________________________________________________________________
_________________________________________________________________________
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.
Visualize S, I, R Trajectories
<- c ("true" = "#440154FF" , "lstm" = "#35B779FF" )<- function (data, state_name, title) {<- data %>% filter (state == state_name)ggplot (plot_data, aes (x = date, color = param_type)) + geom_ribbon (data = plot_data %>% filter (param_type == "lstm" ),aes (ymin = ci_lower, ymax = ci_upper, fill = param_type),alpha = 0.2 , color = NA + geom_line (aes (y = mean_count), linewidth = 1.1 ) + scale_color_manual (values = method_colors) + scale_fill_manual (values = method_colors) + labs (title = title, x = "Day" , y = "Count (95% CI)" , color = "Method" , fill = "Method" ) + theme_minimal () + theme (legend.position = "bottom" , plot.title = element_text (size = 12 , hjust = 0.5 ))<- create_sir_plot (all_simulation_results, "Susceptible" , "Susceptible over Time" )<- create_sir_plot (all_simulation_results, "Infected" , "Infected over Time" )<- create_sir_plot (all_simulation_results, "Recovered" , "Recovered over Time" )/ p_inf / p_rec) + plot_layout (guides = "collect" ) + plot_annotation (title = "SIR Dynamics: True vs LSTM-Calibrated Parameters" ,subtitle = paste0 ("Each method averaged over " , n_reps, " simulations" )
Bias Tables
Parameter Bias
<- tibble (Parameter = c ("Contact Rate" , "Transmission Rate" , "R0" ),True = c (true_params$ contact_rate, true_params$ transmission_rate, true_params$ R0),LSTM = c (lstm_params$ contact_rate, lstm_params$ transmission_rate, lstm_params$ R0)%>% mutate (Bias = LSTM - True,
# A tibble: 3 × 4
Parameter True LSTM Bias
<chr> <dbl> <dbl> <dbl>
1 Contact Rate 1.76 2.47 0.703
2 Transmission Rate 0.149 0.109 -0.0405
3 R0 3.36 3.42 0.0657